Distributed Configuration
OmniSci supports distributed configuration, which allows single queries to span more than one physical host when the scale of the data is too large to fit on a single machine.
In addition to increased capacity, a distributed setup has other advantages. Writes to the database can be distributed across the nodes, thereby speeding up import. Reads from disk are similarly accelerated. For clients that have performance requirements higher than the levels a single node can provide, additional GPUs in a distributed cluster can significantly increase read performance in many usage scenarios. Performance scales linearly, or near linearly, with the number of GPUs, for simple queries requiring little communication between servers.
GPU processing is distributed in a similar fashion. Multiple GPUs across the cluster query data on their local hosts. This allows processing of larger datasets, distributed across multiple servers.
The OmniSci Distributed Cluster
A OmniSci distributed database consists of three components:
- An aggregator, which is a specialized OmniSci Core Database instance for managing the cluster
- One or more leaf nodes, each being a complete OmniSci Core Database instance for storing and querying data
- A String Dictionary Server, which is a centralized repository for all dictionary-encoded items
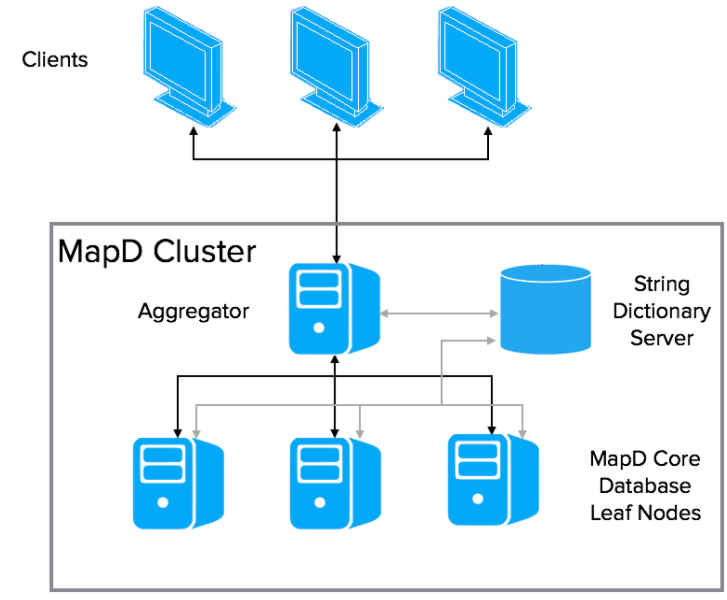
Conceptually, a OmniSci distributed database is horizontally sharded across n leaf nodes. Each leaf node holds one nth of the total dataset. Sharding currently is round-robin only. Queries and responses are orchestrated by a OmniSci Aggregator server.
The OmniSci Aggregator
Clients interact with the aggregator. The aggregator orchestrates execution of a query across the appropriate leaf nodes. The aggregator composes the steps of the query execution plan to send to each leaf node, and manages their results. The full query execution might require multiple iterations between the aggregator and leaf nodes before returning a result to the client.
A core feature of the OmniSci Core Database is backend, GPU-based rendering for data-rich charts such as point maps. When running as a distributed cluster, the backend rendering is distributed across all leaf nodes, and the aggregator composes the final image.
String Dictionary Server
The String Dictionary Server manages and allocates IDs for dictionary-encoded fields, ensuring that these IDs are consistent across the entire cluster.
The server creates a new ID for each new encoded value. For queries returning results from encoded fields, the IDs are automatically converted to the original values by the aggregator. Leaf nodes use the string dictionary for processing joins on encoded columns.
For moderately sized configurations, the String Dictionary Server can share a host with a leaf node. For larger clusters, this service can be configured to run on a small, separate CPU-only server.
Replicated Tables
A table is split by default to 1/nth of the complete dataset. When you create a table used to provide dimension information, you can improve performance by replicating its contents onto every leaf node using the partitions property. For example:
CREATE TABLE flights … WITH (PARTITIONS=’REPLICATED’)
This reduces the distribution overhead during query execution in cases where sharding is not possible or appropriate. This is most useful for relatively small, heavily used dimension tables.
Data Loading
You can load data to a OmniSci distributed cluster using a COPY FROM statement to load data to the aggregator, exactly as with OmniSci single-node processing. The aggregator distributes data evenly across the leaf nodes.
Data Compression
Records transferred between systems in an OmniSci cluster are compressed to improve performance. OmniSci uses the LZ4_HC compressor by default. It is the fastest compressor, but has the lowest compression rate of the available algorithms. The time required to compress each buffer is directly proportional to the final compressed size of the data. A better compression rate will likely require more time to process.
You can specify another compressor on server startup using the runtime flag
compressor. Compressor choices include:
- blosclz
- lz4
- lz4hc
- snappy
- zlib
- zstd
For more information on the compressors used with OmniSci, see also:
- http://blosc.org/pages/synthetic-benchmarks/
- https://quixdb.github.io/squash-benchmark/
- https://lz4.github.io/lz4/
OmniSci does not compress the payload until it reaches a certain size. The
default size limit is 512MB. You can change the size using the runtime flag
compression-limit-bytes.
An Example OmniSci Distributed Cluster
This example uses four GPU-based machines, each with a combination of one or more CPUs and GPUs.
| Hostname | IP | Role(s) |
|---|---|---|
| Node1 | 10.10.10.1 | Leaf, Aggregator |
| Node2 | 10.10.10.2 | Leaf, String Dictionary Server |
| Node3 | 10.10.10.3 | Leaf |
| Node4 | 10.10.10.4 | Leaf |
Install OmniSci server on each node. For larger deployments, you can have the install on a shared drive.
Set up the configuration file for the entire cluster. This file is the same for all nodes.
[
{
"host": "node1",
"port": 19091,
"role": "dbleaf"
},
{
"host": "node2",
"port": 19091,
"role": "dbleaf"
},
{
"host": "node3",
"port": 19091,
"role": "dbleaf"
},
{
"host": "node4",
"port": 19091,
"role": "dbleaf"
},
{
"host": "node2",
"port": 10301,
"role": "string"
}
]
In the cluster.conf file, the location of each leaf node is identified as well as the location of the String Dictionary server.
Here, dbleaf is a leaf node, and string is the String Dictionary Server. The port each node is listening on is also identified. These ports must match the ports configured on the individual server.
Each leaf node requires a mapd.conf configuration file.
port = 19091 http-port = 19090 calcite-port = 19093 data = "<location>/mapd-storage/nodeLocal/data" read-only = false quiet = false string-servers = "<location>/mapd-storage/cluster.conf"
The parameter string-servers identifies the file containing the cluster configuration, to tell the leaf node where the String Dictionary Server is.
The aggregator node requires a slightly different mapd.conf. The file is named mapd-agg.conf in this example.
port = 9091 http-port = 9090 calcite-port = 9093 data = "<location>/mapd-storage/nodeLocalAggregator/data" read-only = false quiet = false num-gpus = 1 cluster = "<location>/mapd-storage/cluster.conf" [web] port = 9092 frontend = "<location>/prod/mapd/frontend"
The parameter cluster tells the OmniSci Core Database instance that it is an aggregator node, and where to find the rest of its cluster.
If your aggregator node is sharing a machine with a leaf node, there might be a conflict on the calcite-port. Consider changing the port number of the aggregator node to another that is not in use.
Implementing a OmniSci Distributed Cluster
Contact OmniSci support for assistance with OmniSci Distributed Cluster implementation.
| Important | When installing a distributed cluster, you must run initdb --skip-geo to avoid the automatic creation of the sample geospatial data table. Otherwise, metadata across the cluster falls out of synchronization and can put the server in an unusable state. |
